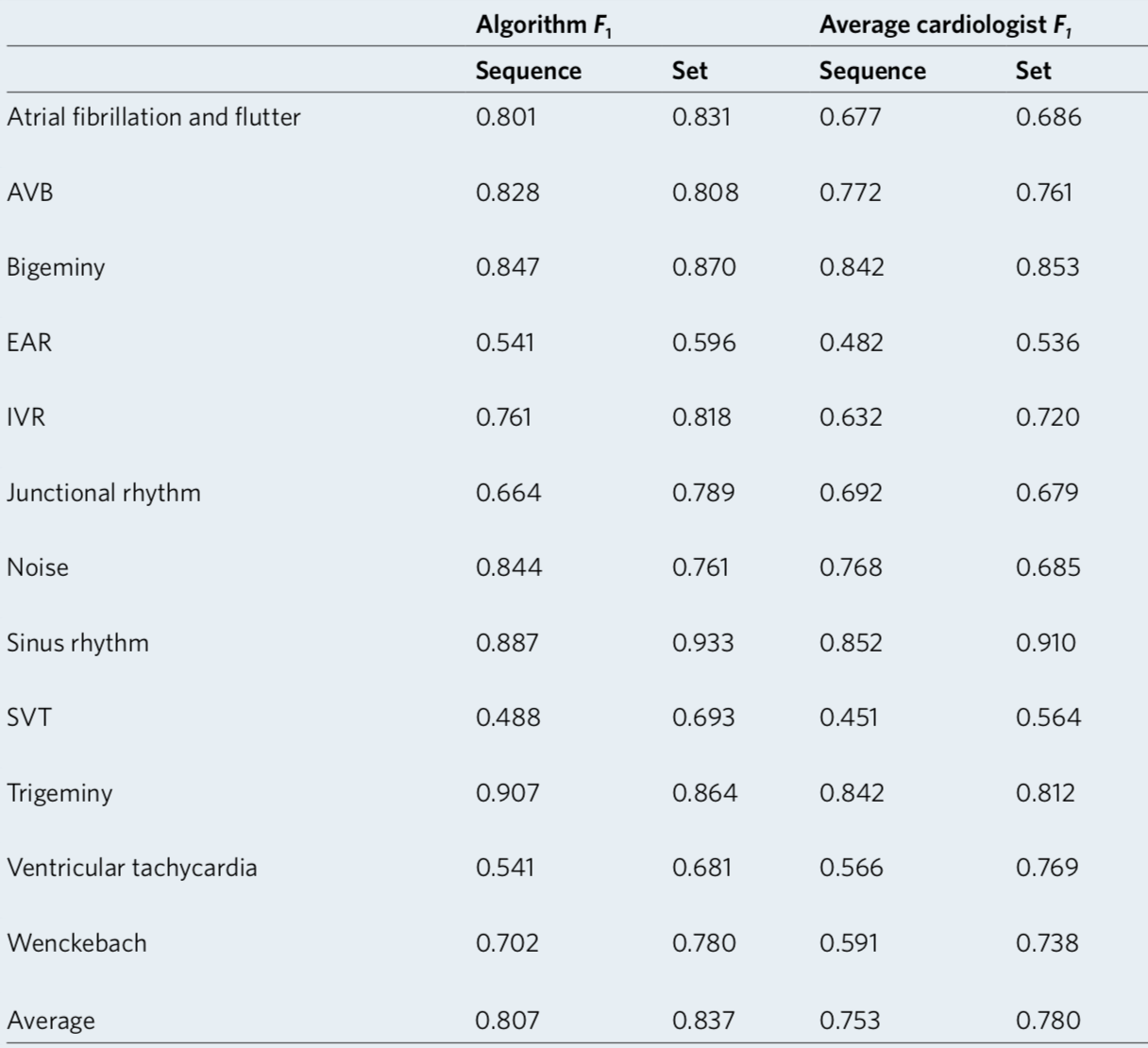
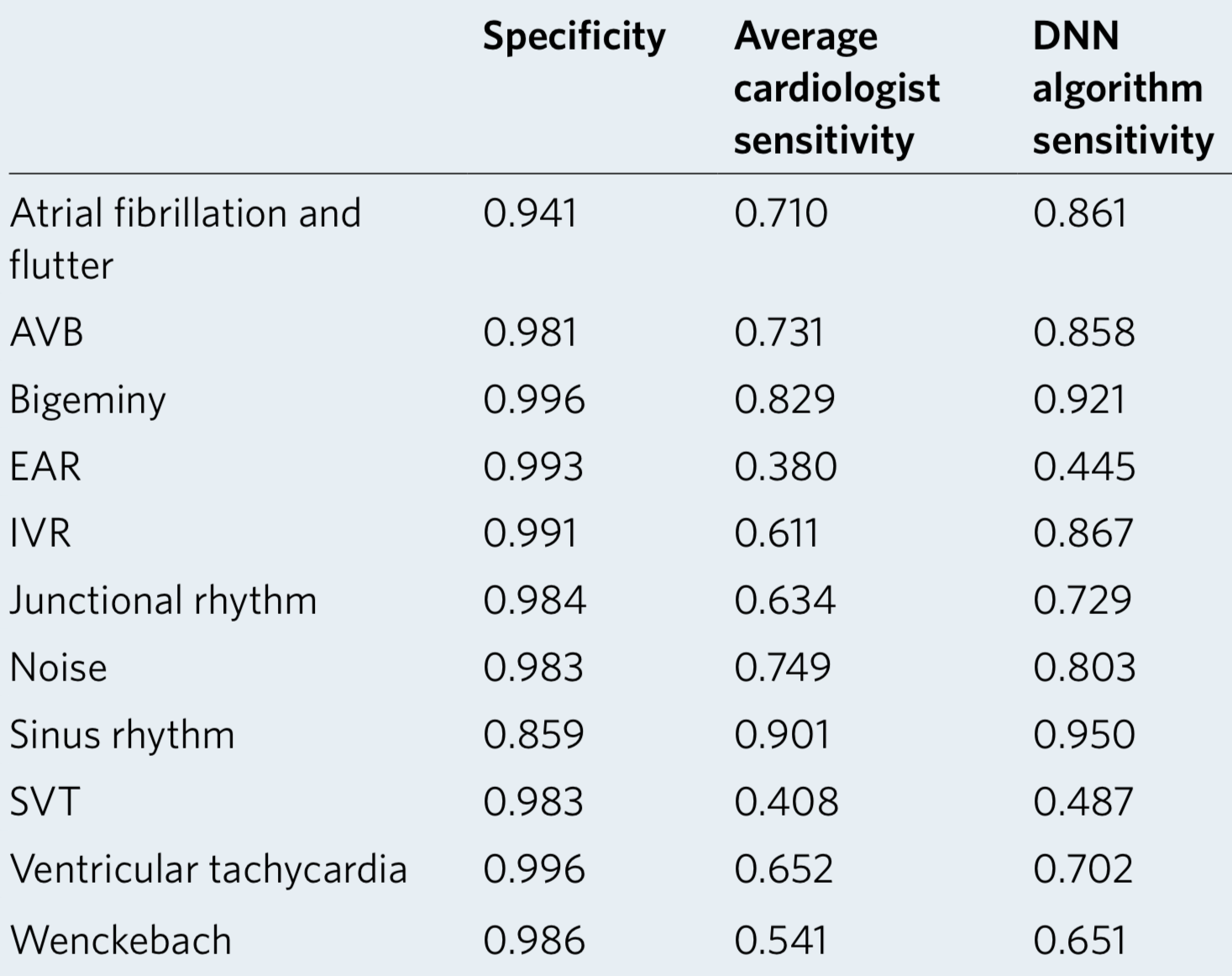
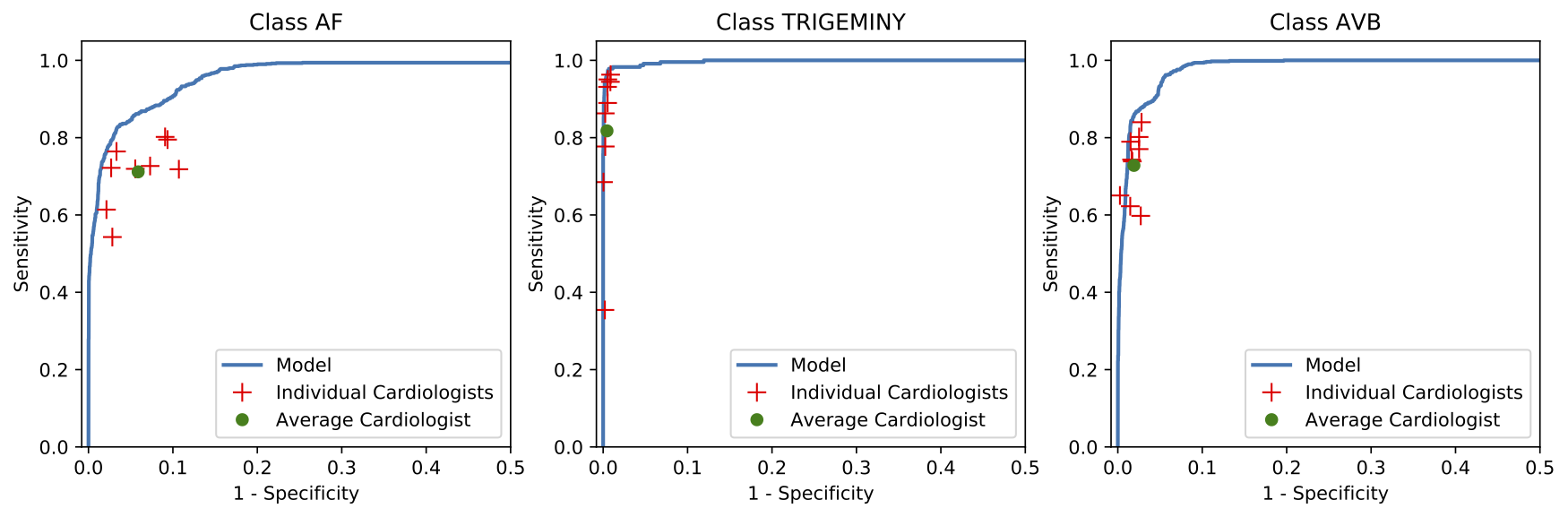
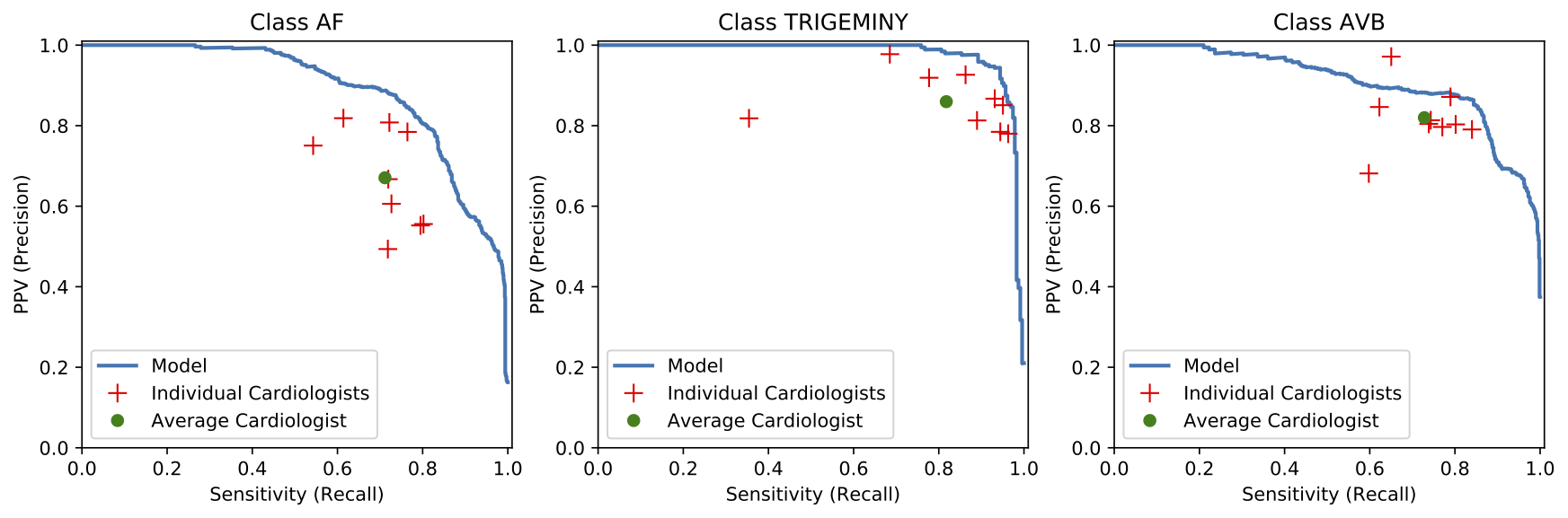
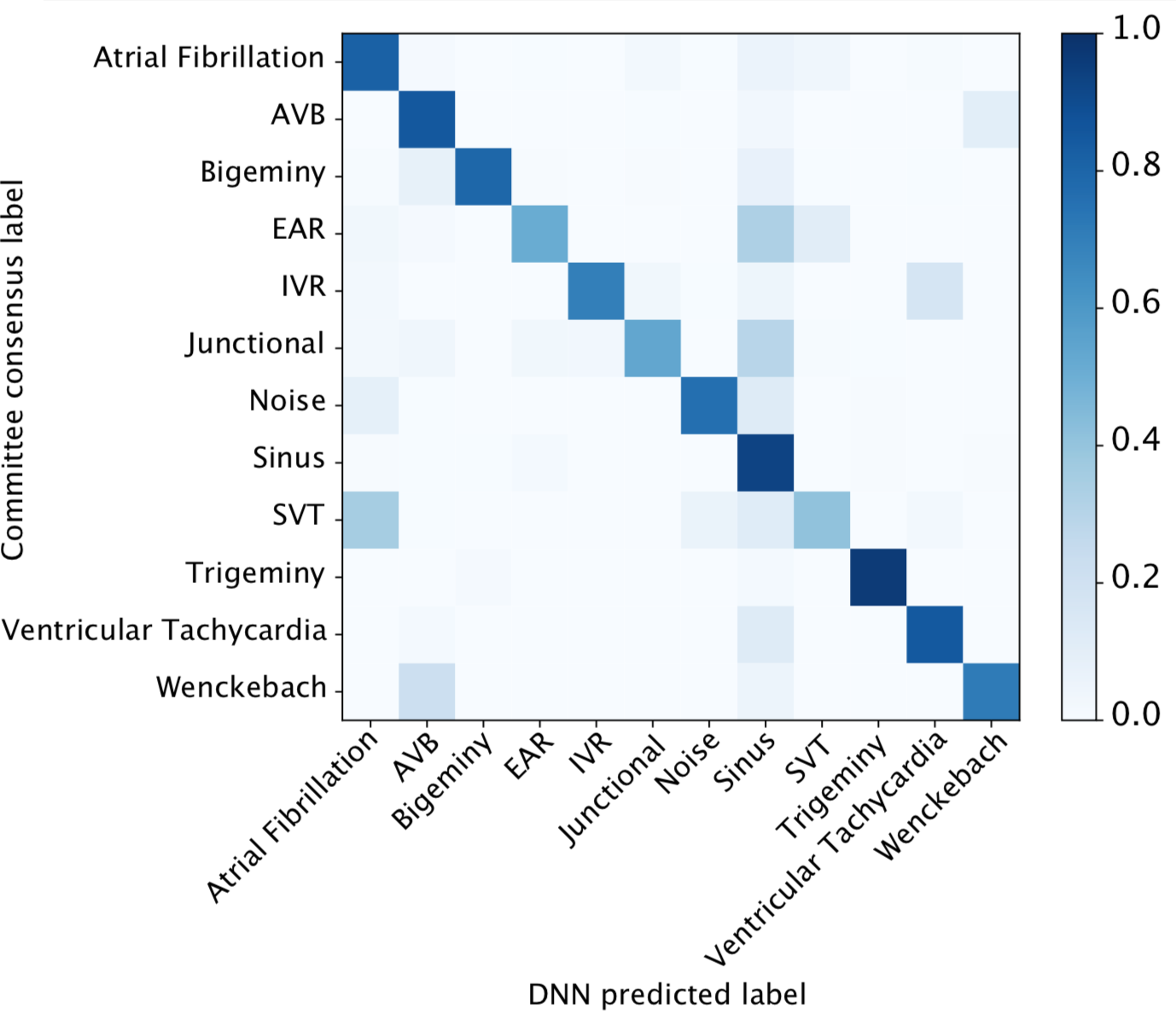
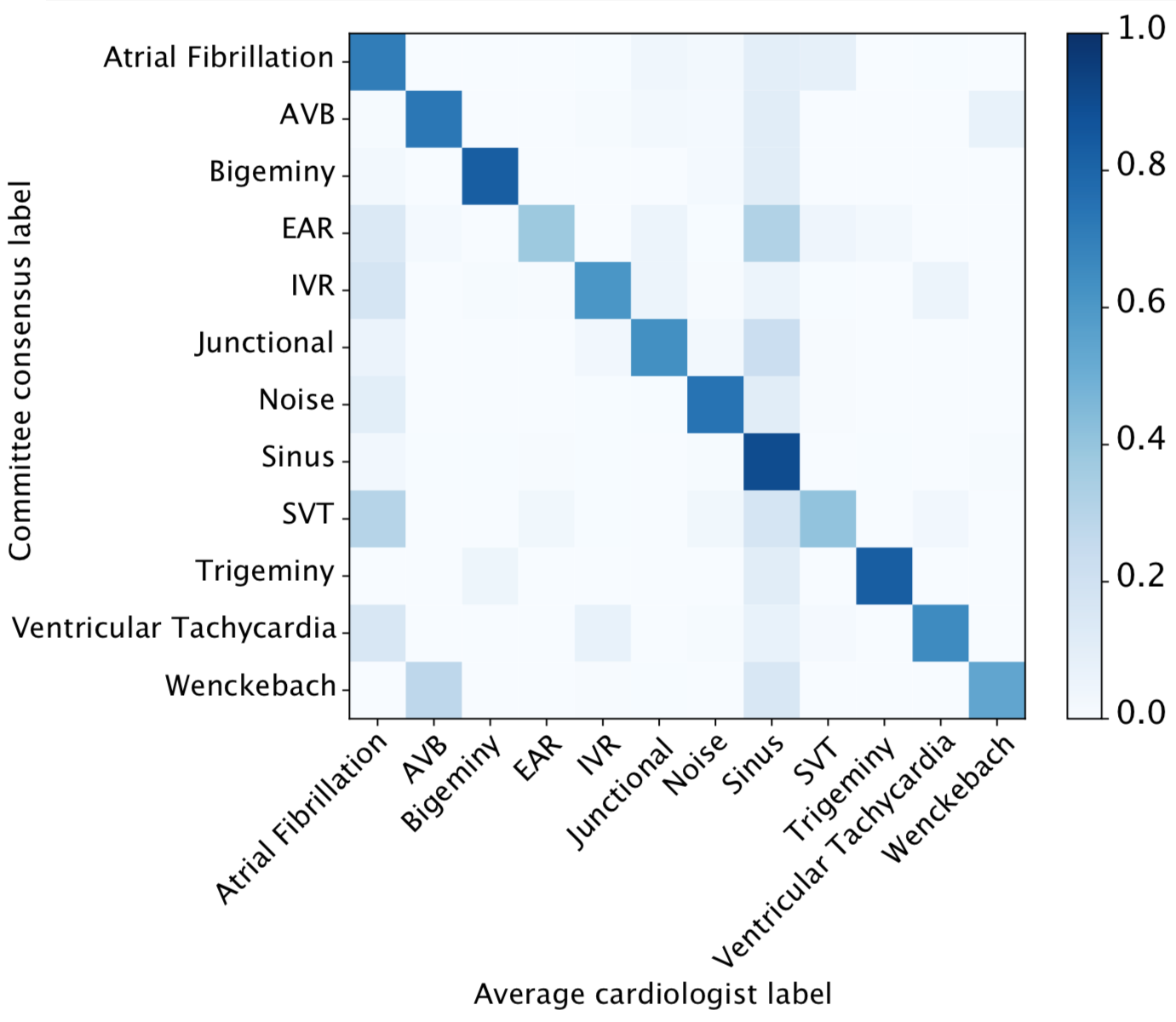
We developed a 1D convolutional deep neural network to detect arrhythmias in arbitrary length ECG time-series.
We developed a convolutional DNN to detect arrhythmias, which takes as input the raw ECG data (sampled at 200 Hz, or 200 samples per second) and outputs one prediction every 256 samples (or every 1.28 s), which we call the output interval. The network takes as input only the raw ECG samples and no other patient- or ECG-related features. The network architecture has 34 layers; to make the optimization of such a network tractable, we employed shortcut connections in a manner similar to the residual network architecture.
Distinct from some other recent DNN approaches, no significant preprocessing of ECG data, such as Fourier or wavelet transforms, is needed to achieve strong classification performance.