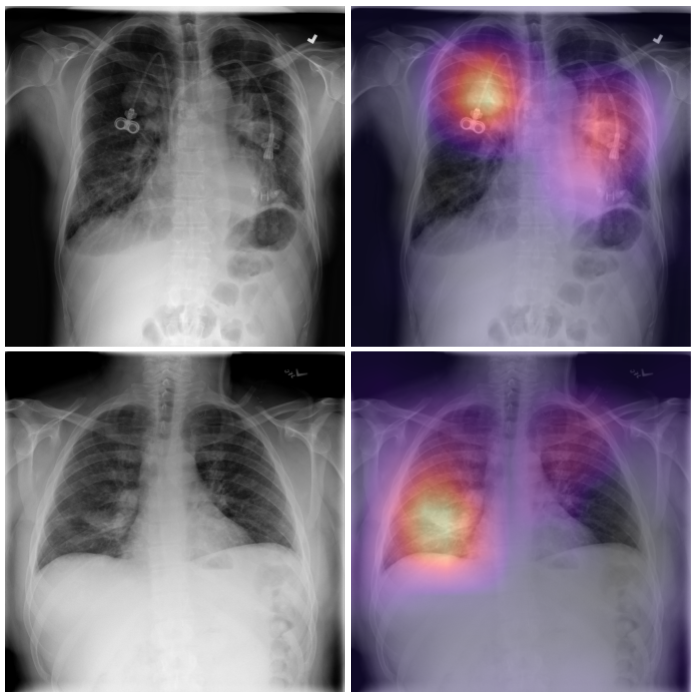
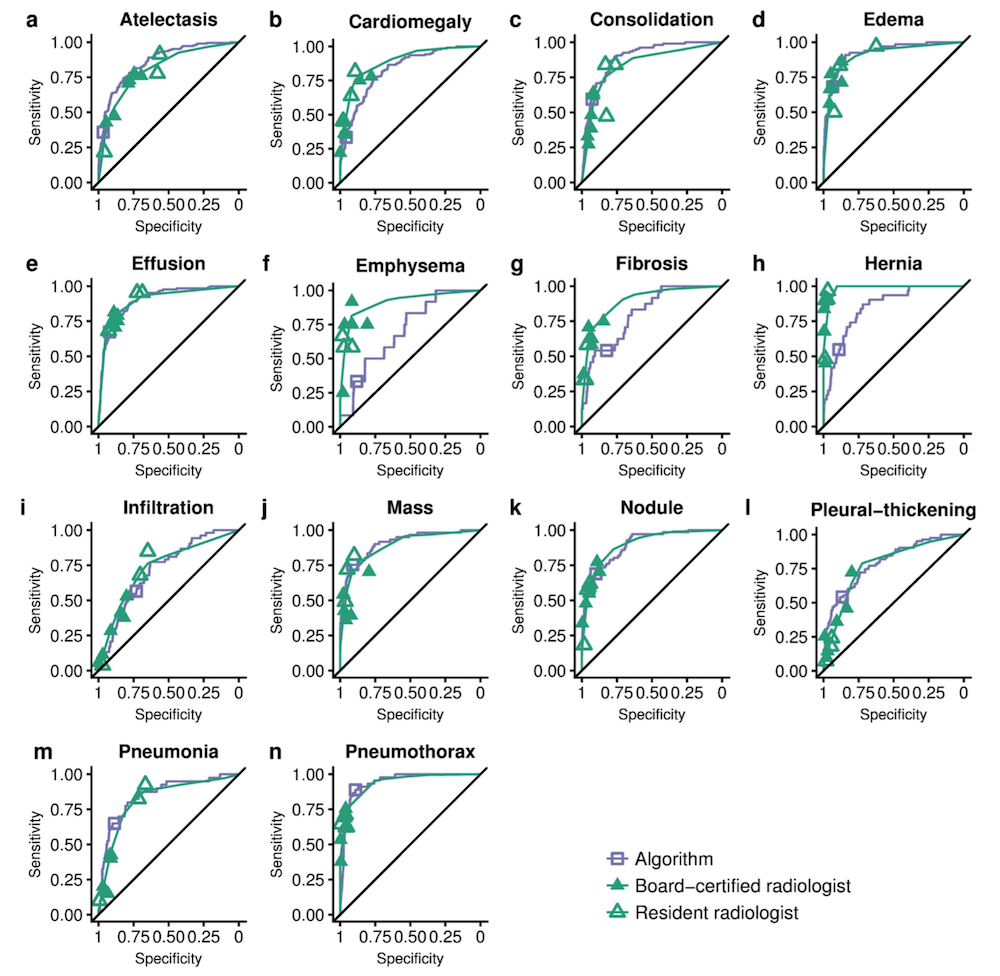
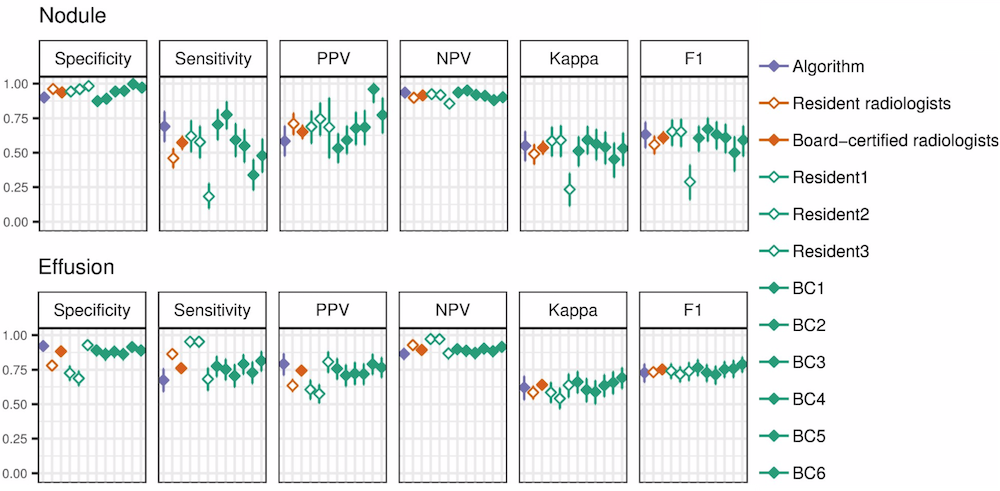
CheXNeXt is trained to predict diseases on x-ray images and highlight parts of an image most indicative of each predicted disease.
CheXNeXt is trained on the ChestX-ray14 dataset, one of the largest public repository of radiographs, containing 112,120 frontal-view chest radiographs of 30,805 unique patients. Each image in ChestX-ray14 was labeled using an automatic extraction method on radiology reports.
CheXNeXt's training process consists of 2 consecutive stages to account for the partially incorrect labels in the ChestX-ray14 dataset. First, an ensemble of networks is trained on the training set to predict the probability that each of the 14 pathologies is present in the image. The predictions of this ensemble are used to relabel the training and tuning sets. A new ensemble of networks are finally trained on this relabeled training set.
Without any additional supervision, CheXNeXt produces heat maps that identify locations in the chest radiograph that contribute most to the network’s classification using class activation mappings (CAMs).