

CheXphoto is a competition for x-ray interpretation based on a new dataset of naturally and synthetically perturbed chest x-rays hosted by Stanford and VinBrain.
Read the Paper (Phillips, Rajpurkar & Sabini et al.)Chest radiography is the most common imaging examination globally, and is critical for screening, diagnosis, and management of many life threatening diseases. Most chest x-ray algorithms have been developed and validated on digital x-rays, while the vast majority of developing regions use films. An appealing solution to scaled deployment is to leverage the ubiquity of smartphones for automated interpretation of film through cellphone photography. Automated interpretation of photos of chest x-rays at the same high-level of performance as with digital chest x-rays is challenging because photographs of x-rays introduce visual artifacts not commonly found in digital x-rays. To encourage high model performance for this application, we developed CheXphoto, a dataset of photos of chest x-rays and synthetic transformations designed to mimic the effects of photography.
With the launch of the CheXphoto competition, we are pleased to announce the release of an additional set of x-ray film images provided by VinBrain, a subsidiary of Vingroup. Please see Validation and Test Sets for details.
Will your model perform as well as radiologists in detecting different pathologies in chest X-rays?
This competition is now closed
| Rank | Date | Model | AUC Film | AUC Digital |
|---|---|---|---|---|
| 1 | Oct 01, 2021 | LBC-v2 (ensemble) Macao Polytechnic Institute | 0.850 | 0.89 |
| 2 | Sep 18, 2021 | LBC-v0 (ensemble) Macao Polytechnic Institute | 0.820 | 0.89 |
| 3 | Aug 26, 2021 | Stellarium-CheXpert-Local (single model) Macao Polytechnic Institute | 0.802 | 0.88 |
| 4 | May 07, 2021 | MVD121 single model | 0.762 | 0.83 |
| 5 | May 11, 2021 | MVD121-320 single model | 0.758 | 0.84 |
| 6 | Jun 22, 2020 | DE_JUN4_RS_EN ensemble LTTS | 0.742 | 0.91 |
| 7 | Aug 11, 2020 | NewTrickTest (ensemble) XBSJ | 0.735 | 0.88 |
| 8 | Jun 29, 2020 | BASELINE Acorn single model | 0.732 | 0.85 |
| 9 | Jun 22, 2020 | DE_JUN2_RS_EN ensemble LTTS | 0.722 | 0.91 |
| 10 | Mar 03, 2021 | mhealth_buet (single model) BUET | 0.710 | 0.87 |
| 11 | Jul 14, 2020 | CombinedTrainDenseNet121 (single model) University of Westminster, Silva R. | 0.707 | 0.86 |
| 12 | Mar 28, 2021 | Yoake (single model) Macao Polytechnic Institute | 0.682 | 0.86 |
| 13 | Jan 12, 2021 | mwowra-conditional (single) AGH UST | 0.646 | 0.80 |
| 14 | Jun 11, 2021 | AccidentNet V2 (single model) Macao Polytechnic Institute | 0.604 | 0.76 |
| 15 | May 25, 2021 | Stellarium (single model) Macao Polytechnic Institute | 0.599 | 0.71 |
| 16 | Sep 24, 2020 | Grp12BigCNN ensemble | 0.595 | 0.82 |
| 17 | Sep 11, 2020 | {koala-large} (single model) SJTU | 0.589 | 0.76 |
| 18 | Aug 17, 2020 | DiseaseNet Samg2003 single model, DPS RKP,http://sambhavgupta.com | 0.583 | 0.83 |
| 19 | Jul 02, 2020 | 12ASLv2(single) AITD | 0.572 | 0.73 |
| 20 | May 28, 2021 | AccidentNet v1 (single model) Macao Polytechnic Institute | 0.561 | 0.74 |
Update: This competition is now closed.
Here's a tutorial walking you through official evaluation of your model. Once your model has been evaluated officially, your scores will be added to the leaderboard.
CheXphoto comprises a training set of natural photos and synthetic transformations of 10,507 x-rays from 3,000 unique patients that were sampled at random from the CheXpert training set, and a validation and test set of natural and synthetic transformations applied to all 234 x-rays from 200 patients and 668 x-rays from 500 patients in the CheXpert validation and test sets, respectively.
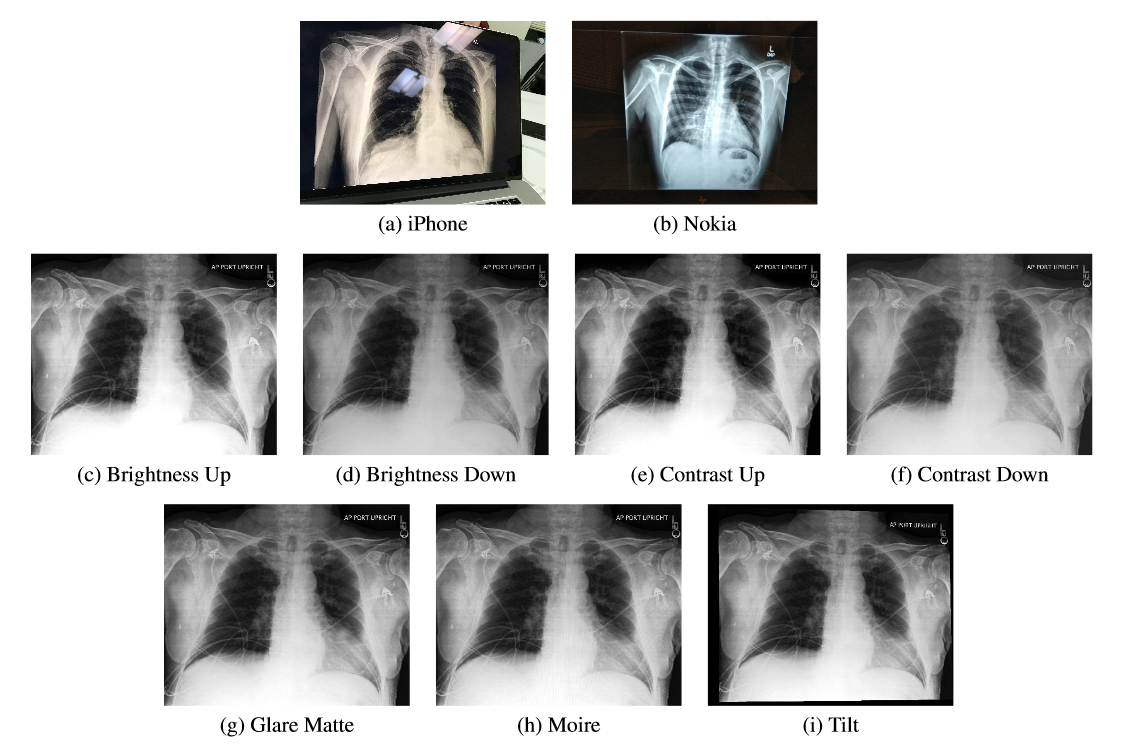
Natural photos consist of x-ray photography using cell phone cameras in various lighting conditions and environments. We developed two sets of natural photos: images captured through an automated process using a Nokia 6.1 cell phone, and images captured manually with an iPhone 8.
Synthetic transformations consist of automatic changes to the digital x-rays designed to make them look like photos of digital x-rays and x-ray films. We developed two sets of complementary synthetic transformations: digital transformations to alter contrast and brightness, and spatial transformations to add glare, moiré effects and perspective changes. To ensure that the level of these transformations did not impact the quality of the image for physician diagnosis, the images were verified by a physician. In some cases, the effects may be visually imperceptible, but may still be adversarial for classification models. For both sets, we apply the transformations to the same set of 10,507 x-rays selected for the Nokia10k dataset.
View on Github
We developed a CheXphoto validation and test set to be used for model validation and evaluation. The validation set comprises natural photos and synthetic transformations of all 234 x-rays in the CheXpert validation set, and is included in the public release, while the test set comprises natural photos of all 668 x-rays in the CheXpert test set, and is withheld for evaluation purposes.
We generated the natural photos of the validation set by manually capturing images of x-rays displayed on a 2560×1080 monitor using a OnePlus 6 cell phone, following a protocol that mirrored the iPhone1k dataset. Synthetic transformations of the validation images were produced using the same protocol as the synthetic training set. The test set was captured using an iPhone 8, following the same protocol as the iPhone1k dataset.
The validation set contains an additional 250 cell phone images of chest x-ray films provided by VinBrain, a subsidiary of Vingroup in Vietnam. These films were originally collected by VinBrain through joint research projects with leading lung hospitals in Vietnam. An additional 250 film images have been withheld in the test set for model evaluation.
Please read the Stanford University School of Medicine CheXphoto Dataset Research Use Agreement. Once you register to download the CheXphoto dataset, you will receive a link to the download over email. Note that you may not share the link to download the dataset with others.
By registering for downloads from the CheXphoto Dataset, you are agreeing to this Research Use Agreement, as well as to the Terms of Use of the Stanford University School of Medicine website as posted and updated periodically at http://www.stanford.edu/site/terms/.
1. Permission is granted to view and use the CheXphoto Dataset without charge for personal, non-commercial research purposes only. Any commercial use, sale, or other monetization is prohibited.
2. Other than the rights granted herein, the Stanford University School of Medicine (“School of Medicine”) retains all rights, title, and interest in the CheXphoto Dataset.
3. You may make a verbatim copy of the CheXphoto Dataset for personal, non-commercial research use as permitted in this Research Use Agreement. If another user within your organization wishes to use the CheXplanation Dataset, they must register as an individual user and comply with all the terms of this Research Use Agreement.
4. YOU MAY NOT DISTRIBUTE, PUBLISH, OR REPRODUCE A COPY of any portion or all of the CheXplanation Dataset to others without specific prior written permission from the School of Medicine.
5. YOU MAY NOT SHARE THE DOWNLOAD LINK to the CheXplanation dataset to others. If another user within your organization wishes to use the CheXplanation Dataset, they must register as an individual user and comply with all the terms of this Research Use Agreement.
6. You must not modify, reverse engineer, decompile, or create derivative works from the CheXplanation Dataset. You must not remove or alter any copyright or other proprietary notices in the CheXplanation Dataset.
7. The CheXplanation Dataset has not been reviewed or approved by the Food and Drug Administration, and is for non-clinical, Research Use Only. In no event shall data or images generated through the use of the CheXplanation Dataset be used or relied upon in the diagnosis or provision of patient care.
8. THE CheXplanation DATASET IS PROVIDED "AS IS," AND STANFORD UNIVERSITY AND ITS COLLABORATORS DO NOT MAKE ANY WARRANTY, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE, NOR DO THEY ASSUME ANY LIABILITY OR RESPONSIBILITY FOR THE USE OF THIS CheXplanation DATASET.
9. You will not make any attempt to re-identify any of the individual data subjects. Re-identification of individuals is strictly prohibited. Any re-identification of any individual data subject shall be immediately reported to the School of Medicine.
10. Any violation of this Research Use Agreement or other impermissible use shall be grounds for immediate termination of use of this CheXplanation Dataset. In the event that the School of Medicine determines that the recipient has violated this Research Use Agreement or other impermissible use has been made, the School of Medicine may direct that the undersigned data recipient immediately return all copies of the CheXplanation Dataset and retain no copies thereof even if you did not cause the violation or impermissible use.
In consideration for your agreement to the terms and conditions contained here, Stanford grants you permission to view and use the CheXplanation Dataset for personal, non-commercial research. You may not otherwise copy, reproduce, retransmit, distribute, publish, commercially exploit or otherwise transfer any material.
You may use CheXplanation Dataset for legal purposes only.
You agree to indemnify and hold Stanford harmless from any claims, losses or damages, including legal fees, arising out of or resulting from your use of the CheXplanation Dataset or your violation or role in violation of these Terms. You agree to fully cooperate in Stanford’s defense against any such claims. These Terms shall be governed by and interpreted in accordance with the laws of California.