

CheXpert is a large dataset of chest X-rays and competition for automated chest x-ray interpretation, which features uncertainty labels and radiologist-labeled reference standard evaluation sets.
Read the Paper (Irvin & Rajpurkar et al.)Chest radiography is the most common imaging examination globally, critical for screening, diagnosis, and management of many life threatening diseases. Automated chest radiograph interpretation at the level of practicing radiologists could provide substantial benefit in many medical settings, from improved workflow prioritization and clinical decision support to large-scale screening and global population health initiatives. For progress in both development and validation of automated algorithms, we realized there was a need for a labeled dataset that (1) was large, (2) had strong reference standards, and (3) provided expert human performance metrics for comparison.
Will your model perform as well as radiologists in detecting different pathologies in chest X-rays?
| Rank | Date | Model | AUC | Num Rads Below Curve |
|---|---|---|---|---|
| 1 | Aug 31, 2020 | DeepAUC-v1 ensemble https://arxiv.org/abs/2012.03173 | 0.930 | 2.8 |
| 2 | Sep 01, 2019 | Hierarchical-Learning-V1 (ensemble) Vingroup Big Data Institute https://arxiv.org/abs/1911.06475 | 0.930 | 2.6 |
| 3 | Oct 15, 2019 | Conditional-Training-LSR ensemble | 0.929 | 2.6 |
| 4 | Dec 04, 2019 | Hierarchical-Learning-V4 (ensemble) Vingroup Big Data Institute https://arxiv.org/abs/1911.06475 | 0.929 | 2.6 |
| 5 | Oct 10, 2019 | YWW(ensemble) JF&NNU https://github.com/jfhealthcare/Chexpert | 0.929 | 2.8 |
| 6 | Oct 17, 2019 | Conditional-Training-LSR-V1 ensemble | 0.929 | 2.6 |
| 7 | Nov 17, 2019 | Hierarchical-Learning-V0 (ensemble) Vingroup Big Data Institute | 0.929 | 2.6 |
| 8 | Sep 09, 2019 | Multi-Stage-Learning-CNN-V3 (ensemble) VINBDI Medical Imaging Team | 0.928 | 2.6 |
| 9 | Dec 30, 2019 | DeepCNNsGM(ensemble) HUST | 0.928 | 2.6 |
| 10 | Dec 30, 2019 | DeepCNNs(ensemble) HUST | 0.927 | 2.6 |
| 11 | Dec 16, 2019 | desmond https://github.com/inisis/chexpert https://github.com/inisis/chexpert) | 0.927 | 3.0 |
| 11 | Dec 23, 2019 | inisis https://github.com/inisis/chexpert https://github.com/inisis/chexpert) | 0.927 | 3.0 |
| 12 | Sep 19, 2019 | SenseXDR ensemble | 0.927 | 2.6 |
| 13 | Sep 18, 2019 | ihil (ensemble) UESTC | 0.927 | 2.6 |
| 14 | Jul 01, 2022 | Anatomy-XNet-V1 ensemble https://arxiv.org/abs/2106.05915 | 0.926 | 2.6 |
| 15 | Jul 31, 2019 | JF aboy ensemble_V2 JF HEALTHCAREhttps://github.com/deadpoppy/CheXpert-Challenge | 0.926 | 3.0 |
| 16 | Sep 01, 2019 | yww | 0.926 | 2.6 |
| 17 | Sep 16, 2019 | DRNet (ensemble) UESTC and SenseTime | 0.926 | 2.6 |
| 18 | Feb 11, 2020 | alimebkovkz | 0.925 | 2.4 |
| 18 | Dec 26, 2019 | hoanganh_VB_ensemble37 | 0.925 | 2.4 |
| 19 | Dec 26, 2019 | hoanganh_VB_ensemble35 | 0.925 | 2.4 |
| 20 | Dec 12, 2019 | Hoang_VB_ensemble31_v1 | 0.924 | 2.4 |
| 21 | Dec 17, 2019 | tedtta | 0.924 | 2.4 |
| 22 | Sep 04, 2019 | uestc | 0.924 | 2.6 |
| 23 | Dec 09, 2019 | Hoang_VB_ensemble31_v2 | 0.924 | 2.4 |
| 24 | Dec 04, 2019 | as-hust-v3 ensemble | 0.924 | 2.4 |
| 25 | Jan 10, 2020 | hoanganh_VB_VN3 | 0.924 | 2.4 |
| 26 | Sep 14, 2019 | Hierarchical-CNN-Ensemble-V1 (ensemble) Vingroup Big Data Institute | 0.924 | 2.4 |
| 27 | Apr 25, 2020 | DE_APR ensemble ltts | 0.923 | 2.6 |
| 28 | Apr 25, 2020 | DE_APR_N ensemble ltts | 0.923 | 2.6 |
| 29 | Dec 10, 2019 | hoanganhcnu_ensemble27_v2 | 0.923 | 2.4 |
| 30 | Aug 22, 2019 | Multi-Stage-Learning-CNN-V2 (ensemble) VINBDI Medical Imaging Team | 0.923 | 2.6 |
| 31 | Dec 16, 2019 | Weighted-CNN(ensemble) HUST | 0.923 | 2.6 |
| 32 | Dec 10, 2019 | hoanganhcnu_ensemble27_v1 | 0.923 | 2.4 |
| 33 | Aug 17, 2019 | YJ&&YWW :https://github.com/inisis/chexpert | 0.923 | 2.4 |
| 34 | Sep 15, 2022 | Maxium (ensemble) Macao Polytechnic University | 0.923 | 2.4 |
| 35 | Dec 04, 2019 | as-hust-v1 ensemble | 0.923 | 2.4 |
| 36 | Dec 16, 2019 | Average-CNN(ensemble) HUST | 0.922 | 2.4 |
| 37 | Dec 04, 2019 | as-hust-v2 ensemble | 0.922 | 2.8 |
| 38 | Aug 04, 2020 | MaxAUC ensemble | 0.922 | 2.4 |
| 39 | Nov 21, 2019 | hoangnguyenkcv17 | 0.921 | 2.4 |
| 40 | Sep 04, 2019 | null | 0.922 | 2.2 |
| 41 | Sep 02, 2020 | SuperCNNv3 ensemble | 0.921 | 2.4 |
| 42 | Aug 13, 2019 | null | 0.921 | 2.2 |
| 43 | Aug 15, 2019 | zjr(ensembel) CSU | 0.921 | 2.6 |
| 44 | Aug 15, 2019 | hyc ensemble | 0.921 | 2.4 |
| 45 | Jan 10, 2020 | HOANG_VB_VN_2 ensemble | 0.920 | 2.4 |
| 46 | Nov 23, 2019 | null | 0.920 | 2.6 |
| 47 | Aug 18, 2019 | BDNB ensemble | 0.919 | 2.6 |
| 48 | Dec 20, 2019 | thang ensemble coloa | 0.919 | 2.4 |
| 49 | Nov 30, 2019 | null | 0.919 | 2.2 |
| 50 | Jul 16, 2019 | JF Coolver ensemble ensemble model | 0.919 | 2.6 |
| 51 | Nov 21, 2019 | hoangnn9 ensemble VBVN | 0.919 | 2.4 |
| 52 | Jul 27, 2019 | JF aboy ensemble_V1 JF HEALTHCAREhttps://github.com/deadpoppy/CheXpert-Challenge | 0.919 | 2.4 |
| 53 | Sep 08, 2022 | A Good Model (single model) Macao Polytechnic University | 0.918 | 2.6 |
| 54 | Nov 07, 2019 | brian-baseline-v2 ensemble | 0.919 | 2.2 |
| 55 | Jun 22, 2020 | DE_JUN4_RS_EN ensemble LTTS | 0.918 | 2.6 |
| 56 | Jun 22, 2019 | Mehdi_You (ensemble) IPM_HPC | 0.918 | 2.6 |
| 57 | Apr 17, 2022 | Anatomy-XNet (ensemble) mHealth-BUET https://arxiv.org/abs/2106.05915 | 0.917 | 2.6 |
| 58 | Oct 01, 2021 | Overfit ensemble OTH-AW | 0.917 | 2.2 |
| 59 | Aug 15, 2019 | Deep-CNNs-V1 ensemble | 0.917 | 2.2 |
| 60 | Nov 22, 2019 | thangbk(ensemble) SNU | 0.917 | 2.0 |
| 61 | Jul 18, 2019 | Ensemble_v2 Ian, Wingspan https://github.com/Ien001/CheXpert_challenge_2019 | 0.917 | 2.4 |
| 62 | Jan 21, 2020 | vdn6 ensemble ltts | 0.917 | 2.2 |
| 63 | Jun 22, 2020 | DE_JUN3_RS_EN ensemble LTTS | 0.916 | 2.4 |
| 64 | Nov 25, 2019 | null | 0.916 | 2.4 |
| 65 | Nov 25, 2019 | ATT-AW-v1 ensemble | 0.916 | 2.4 |
| 66 | Nov 14, 2019 | null | 0.916 | 2.2 |
| 67 | Dec 14, 2019 | desmond https://github.com/inisis/chexpert https://github.com/inisis/chexpert) | 0.916 | 2.6 |
| 67 | Jun 22, 2020 | DE_JUN1_RS_EN ensemble LTTS | 0.916 | 2.6 |
| 68 | Oct 10, 2019 | desmond https://github.com/inisis/chexpert https://github.com/inisis/chexpert) | 0.916 | 2.6 |
| 69 | Aug 25, 2019 | Multi-Stage-Learning-CNN-V0 ensemble | 0.916 | 2.2 |
| 70 | Aug 19, 2019 | TGNB ensemble | 0.915 | 2.6 |
| 71 | Jul 15, 2019 | Deadpoppy Ensemble ensemble model | 0.915 | 2.2 |
| 72 | Nov 18, 2019 | hoangnguyenkcv-ensemble28 ensemble | 0.915 | 2.2 |
| 73 | Aug 05, 2019 | zhangjingyang | 0.915 | 2.4 |
| 74 | Dec 10, 2019 | ensemble SNU | 0.915 | 2.4 |
| 75 | Jun 22, 2020 | DE_JUN2_RS_EN ensemble LTTS | 0.914 | 2.6 |
| 76 | Aug 16, 2019 | GRNB ensemble | 0.914 | 2.4 |
| 77 | Jul 31, 2019 | Deep-CNNs (ensemble) Vingroup Big Data Institute | 0.914 | 2.0 |
| 78 | Dec 02, 2019 | Sky-Model ensemble | 0.913 | 2.2 |
| 79 | Jul 23, 2019 | JF Deadpoppy ensemble | 0.913 | 2.2 |
| 80 | Aug 14, 2019 | YWW-YJ:https://github.com/inisis/chexpert | 0.913 | 2.0 |
| 81 | Aug 17, 2019 | zjy ensemble | 0.912 | 2.2 |
| 82 | Jun 29, 2020 | WL_Baseline (ensemble) WL | 0.912 | 2.0 |
| 83 | Oct 30, 2021 | anatomy_xnet_v1 (single model) BUET | 0.911 | 2.2 |
| 84 | Aug 01, 2019 | songtao | 0.911 | 2.2 |
| 85 | Oct 25, 2019 | bhtrung | 0.911 | 2.2 |
| 85 | Oct 27, 2019 | KCV-CNN-ensemble-CNU | 0.911 | 2.2 |
| 86 | Apr 25, 2020 | DS_APR_N single model ltts | 0.911 | 2.0 |
| 87 | Apr 25, 2020 | DS_APR single model LTTS | 0.911 | 2.0 |
| 88 | Oct 24, 2019 | brian-baseline ensemble | 0.911 | 2.0 |
| 89 | Dec 10, 2019 | ensemble SNU3 | 0.910 | 2.2 |
| 90 | Dec 09, 2019 | HinaNetV2 (ensemble) VietAI http://vietai.org | 0.909 | 2.2 |
| 91 | Oct 21, 2022 | KD-Prune10 (Single model) MPU | 0.909 | 2.0 |
| 92 | Jun 05, 2022 | G_Mans_ensemble | 0.909 | 1.8 |
| 93 | Jan 12, 2020 | vdnnn (ensemble) LTTS | 0.908 | 1.8 |
| 94 | May 25, 2022 | BAAZT Ensemble | 0.908 | 1.8 |
| 95 | Jul 31, 2019 | guran_rib | 0.908 | 2.0 |
| 96 | Jan 12, 2020 | vbn (single model) LTTS | 0.907 | 1.6 |
| 97 | Apr 06, 2019 | muti_base (ensemble) SCU_MILAB | 0.907 | 1.6 |
| 98 | Jan 23, 2019 | Stanford Baseline (ensemble) Stanford University https://arxiv.org/abs/1901.07031 | 0.907 | 1.8 |
| 99 | Mar 02, 2022 | Z_Ensemble_V1 | 0.907 | 1.4 |
| 100 | Sep 11, 2019 | {ForwardModelEnsembleCorrected} (ensemble) Stanford | 0.906 | 1.6 |
| 101 | Jun 14, 2019 | Multi-CNN ensemble | 0.905 | 2.4 |
| 102 | Oct 13, 2021 | LBC-v2 (ensemble) Macao Polytechnic University https://arxiv.org/abs/2210.05954 | 0.906 | 1.6 |
| 103 | Jul 25, 2019 | hyc | 0.905 | 1.8 |
| 104 | Oct 02, 2019 | ForwardMECorrectedFull (ensemble) Institution | 0.905 | 1.6 |
| 105 | Jul 21, 2019 | Multi-CNN ensemble | 0.905 | 2.0 |
| 106 | Jun 22, 2019 | JustAnotherDensenet single model | 0.904 | 1.2 |
| 107 | May 06, 2022 | Orlando (single model) Macao Polytechnic University | 0.903 | 1.6 |
| 108 | May 30, 2022 | Max (single model) Macao Polytechnic University | 0.902 | 2.0 |
| 109 | Mar 03, 2020 | DeepLungsEnsemble Alimbekov R. & Vassilenko I. | 0.902 | 1.8 |
| 110 | Sep 29, 2019 | Nakajima_ayase | 0.901 | 1.4 |
| 111 | Jul 04, 2019 | Ensemble_v1 Ian, Wingspan https://github.com/Ien001/CheXpert_challenge_2019 | 0.901 | 1.6 |
| 112 | May 24, 2019 | MLC11 NotDense (single-model) Leibniz University Hannover | 0.900 | 1.6 |
| 113 | Jan 23, 2020 | vn_2 single_model ltts | 0.900 | 1.2 |
| 114 | Apr 19, 2022 | Z_Ensemble_2 | 0.899 | 1.8 |
| 115 | Jul 22, 2019 | adoudou | 0.899 | 1.6 |
| 116 | Jul 22, 2019 | {AVG_MAX}(ensemble) NNU | 0.899 | 2.0 |
| 117 | Aug 01, 2019 | null | 0.899 | 1.6 |
| 118 | Jul 24, 2019 | llllldz single model | 0.899 | 1.6 |
| 118 | Aug 17, 2020 | DiseaseNet Samg2003 single model, DPS RKP,http://sambhavgupta.com | 0.899 | 1.6 |
| 119 | Sep 18, 2021 | LBC-v0 (ensemble) Macao Polytechnic University https://arxiv.org/abs/2210.05954 | 0.899 | 1.4 |
| 120 | Nov 09, 2019 | BUAA | 0.898 | 1.8 |
| 121 | Jan 29, 2022 | G_Mans_v2 (single model): LibAUC + coat_mini timm lib | 0.898 | 1.4 |
| 122 | May 07, 2021 | ljc2266 | 0.898 | 1.2 |
| 123 | Jun 03, 2019 | ForwardModelEnsemble (ensemble) Stanford | 0.897 | 1.6 |
| 124 | Aug 11, 2020 | NewTrickTest (ensemble) XBSJ | 0.897 | 1.6 |
| 125 | May 28, 2021 | AccidentNet v1 (single model) Macao Polytechnic Institute | 0.897 | 1.2 |
| 126 | Feb 05, 2020 | ylz-v01 single model | 0.896 | 1.6 |
| 127 | Aug 26, 2021 | Stellarium-CheXpert-Local single model https://arxiv.org/abs/2210.05954 | 0.896 | 1.4 |
| 128 | Jun 25, 2019 | ldz single model | 0.896 | 1.4 |
| 129 | Feb 17, 2020 | Densenet single | 0.896 | 1.4 |
| 130 | Jul 13, 2019 | Deadpoppy Single single model | 0.895 | 1.8 |
| 131 | Aug 02, 2019 | adoudou | 0.895 | 1.6 |
| 132 | Sep 11, 2020 | {koala-large} (single model) SJTU | 0.895 | 1.4 |
| 133 | Dec 26, 2019 | MM1 ensemble | 0.894 | 1.6 |
| 134 | May 07, 2021 | MVD121 single model | 0.895 | 1.2 |
| 135 | Jul 21, 2019 | hust(single model) HUST | 0.895 | 1.0 |
| 136 | Jul 29, 2019 | zhujieru | 0.894 | 1.6 |
| 136 | Jul 28, 2019 | hycNB | 0.894 | 1.6 |
| 137 | Jul 04, 2019 | U-Random-Ind (single) BHSB | 0.894 | 1.0 |
| 138 | Jun 03, 2019 | HybridModelEnsemble (ensemble) Stanford | 0.892 | 1.6 |
| 139 | May 11, 2021 | MVD121-320 single model | 0.891 | 1.2 |
| 140 | Feb 05, 2020 | ylz-v02 single model | 0.891 | 1.0 |
| 141 | Jul 31, 2019 | pause single model | 0.890 | 1.0 |
| 142 | Jul 22, 2019 | Haruka_Hamasaki | 0.890 | 0.80 |
| 142 | Aug 27, 2019 | Haruka_Hamasaki | 0.890 | 0.80 |
| 143 | Aug 05, 2019 | DenseNet169 at 320x320 (single model) Lafayette | 0.889 | 1.4 |
| 144 | Apr 12, 2020 | LR-baseline (ensemble) IITB | 0.889 | 1.4 |
| 145 | Jul 05, 2019 | DataAugFTW (single model) University Hannover | 0.888 | 1.0 |
| 146 | Aug 25, 2020 | {koala} (single model) SJTU | 0.888 | 1.0 |
| 147 | Nov 02, 2022 | pm_rn50_0.15ppl | 0.887 | 1.2 |
| 148 | May 25, 2021 | Stellarium single model | 0.887 | 1.2 |
| 149 | Nov 01, 2022 | PrateekMunjal | 0.886 | 1.0 |
| 150 | Oct 22, 2019 | baseline3 single model | 0.886 | 1.2 |
| 151 | May 07, 2021 | MVR50 single model | 0.886 | 0.80 |
| 152 | Aug 25, 2022 | MNet-Fix (Single Model) MPU | 0.884 | 1.6 |
| 153 | Jun 04, 2019 | Coolver XH single model | 0.884 | 0.80 |
| 154 | Mar 24, 2019 | Naive Densenet single model https://github.com/simongrest/chexpert-entries | 0.883 | 1.2 |
| 155 | Mar 03, 2021 | mhealth_buet (single model) BUET | 0.883 | 0.60 |
| 156 | Apr 17, 2021 | Aoitori (single model) Macao Polytechnic Institute | 0.882 | 0.80 |
| 157 | Feb 01, 2022 | {chexpert-classifier}(single model) G42 | 0.882 | 0.60 |
| 158 | Mar 01, 2021 | DearBrave (single model) Macao Polytechnic Institute | 0.882 | 0.40 |
| 159 | Jun 11, 2021 | AccidentNet V2 (single model) Macao Polytechnic Institute | 0.881 | 1.0 |
| 160 | May 04, 2019 | {densenet} (single model) Microsoft | 0.880 | 1.2 |
| 161 | Mar 28, 2021 | Yoake (single model) Macao Polytechnic Institute | 0.879 | 0.60 |
| 162 | May 14, 2019 | MLC11 Baseline (single-model) Leibniz University Hannover | 0.878 | 0.60 |
| 163 | May 27, 2019 | null | 0.878 | 0.80 |
| 164 | Jul 07, 2019 | DenseNet single | 0.876 | 1.2 |
| 165 | Nov 22, 2019 | HCL1 (single model) LTTS | 0.876 | 1.0 |
| 166 | Oct 28, 2019 | GCN_densenet121-single model | 0.875 | 1.0 |
| 167 | Jul 11, 2019 | MLGCN (single model) sensetime | 0.875 | 1.2 |
| 168 | Feb 22, 2021 | GreenTeaCalpis (single model) Macao Polytechnic Institute | 0.873 | 0.80 |
| 169 | Jun 03, 2019 | Multi-CNN (ensemble) VinGroup Big Data Institute | 0.873 | 0.40 |
| 170 | Oct 25, 2021 | BASELINE ResNet50 ensemble | 0.871 | 0.60 |
| 171 | Aug 16, 2021 | Baseline DenseNet161 single model http://www.cadcovid19.dcc.ufmg.br/classification | 0.868 | 0.60 |
| 172 | Oct 14, 2019 | baseline1 (single model) Endimension | 0.868 | 0.80 |
| 173 | May 03, 2020 | DSENet single model | 0.865 | 0.60 |
| 174 | Apr 26, 2019 | Densenet-Basic Single NUST | 0.863 | 0.80 |
| 175 | Aug 12, 2022 | KD_Mobilenet (single model) IPM | 0.862 | 0.80 |
| 176 | May 27, 2019 | null | 0.862 | 0.40 |
| 177 | Apr 25, 2019 | {GoDense} (single model) UPenn | 0.861 | 1.0 |
| 178 | May 16, 2019 | inceptionv3_single_NNU | 0.861 | 0.40 |
| 179 | Aug 22, 2022 | MLKD (Single model) IPM | 0.860 | 0.80 |
| 180 | Jun 29, 2020 | BASELINE Acorn single model | 0.860 | 0.60 |
| 181 | Mar 05, 2022 | ErrorNet (single model) IPM | 0.859 | 0.60 |
| 181 | Mar 18, 2022 | SleepNet (single model) MPI | 0.859 | 0.60 |
| 182 | Oct 22, 2019 | baseline2 single model | 0.858 | 1.0 |
| 183 | Oct 04, 2022 | UMLS_CLIP (single model) SJTU | 0.858 | 0.0 |
| 184 | Feb 27, 2022 | haw02 (single model) IPM | 0.854 | 0.80 |
| 185 | Jul 14, 2020 | CombinedTrainDenseNet121 (single model) University of Westminster, Silva R. | 0.853 | 0.0 |
| 186 | Apr 28, 2019 | rayOfLightSingle (Single Model) GeorgiaTech CSE6250 Team58 | 0.851 | 0.40 |
| 187 | Apr 24, 2019 | Model_Team_34 (single model) Gatech | 0.850 | 0.60 |
| 188 | Apr 27, 2019 | Test model habbes | 0.850 | 0.40 |
| 189 | Feb 02, 2021 | model2_DenseNet121 single | 0.848 | 0.60 |
| 190 | Apr 27, 2019 | Baseline Ensemble | 0.848 | 0.20 |
| 191 | Nov 01, 2019 | HinaNet (single model) VietAI http://vietai.org | 0.844 | 0.40 |
| 192 | Apr 24, 2019 | singlehead_models (single model combined) Gatech CSE6250 Team30 | 0.842 | 0.20 |
| 193 | Jan 12, 2021 | mwowra-conditional (single) AGH UST | 0.840 | 0.40 |
| 194 | Apr 24, 2019 | multihead_model (one model for all pathologies) Gatech CSE6250 Team30 | 0.838 | 0.40 |
| 195 | Aug 12, 2022 | mobilenet (single model) ipm | 0.837 | 0.20 |
| 196 | Jun 13, 2019 | MLC9_Densenet (single model) Leibniz University Hannover | 0.834 | 0.40 |
| 197 | Sep 24, 2020 | Grp12BigCNN ensemble | 0.835 | 0.0 |
| 198 | Feb 16, 2020 | Grp12v2USup2OSamp (ensemble) AITD | 0.830 | 0.20 |
| 199 | Mar 03, 2020 | null | 0.829 | 0.20 |
| 200 | May 16, 2019 | DNET121-single Ian,Wingspan http://www.wingspan.cn/ | 0.822 | 0.0 |
| 201 | Feb 03, 2020 | null | 0.821 | 0.40 |
| 202 | Jul 02, 2020 | 12ASLv2(single) AITD | 0.769 | 0.0 |
| 203 | May 11, 2019 | DenseNet121 (single model) hemil10 | 0.760 | 0.0 |
| 204 | Jul 02, 2020 | 12ASLv1(single) AITD | 0.736 | 0.0 |
| 205 | Jun 23, 2020 | null | 0.732 | 0.0 |
| 206 | Apr 26, 2019 | rayOfLight (ensemble) GeorgiaTech CSE6250 Team58 | 0.727 | 0.0 |
| 207 | Nov 25, 2019 | BASELINE DenseNet121 single model | 0.724 | 0.0 |
| 208 | Jun 23, 2020 | null | 0.701 | 0.0 |
Update: the competition is now closed.
CheXpert is a large public dataset for chest radiograph interpretation, consisting of 224,316 chest radiographs of 65,240 patients. We retrospectively collected the chest radiographic examinations from Stanford Hospital, performed between October 2002 and July 2017 in both inpatient and outpatient centers, along with their associated radiology reports.
Each report was labeled for the presence of 14 observations as positive, negative, or uncertain. We decided on the 14 observations based on the prevalence in the reports and clinical relevance, conforming to the Fleischner Society’s recommended glossary whenever applicable. We then developed an automated rule-based labeler to extract observations from the free text radiology reports to be used as structured labels for the images.
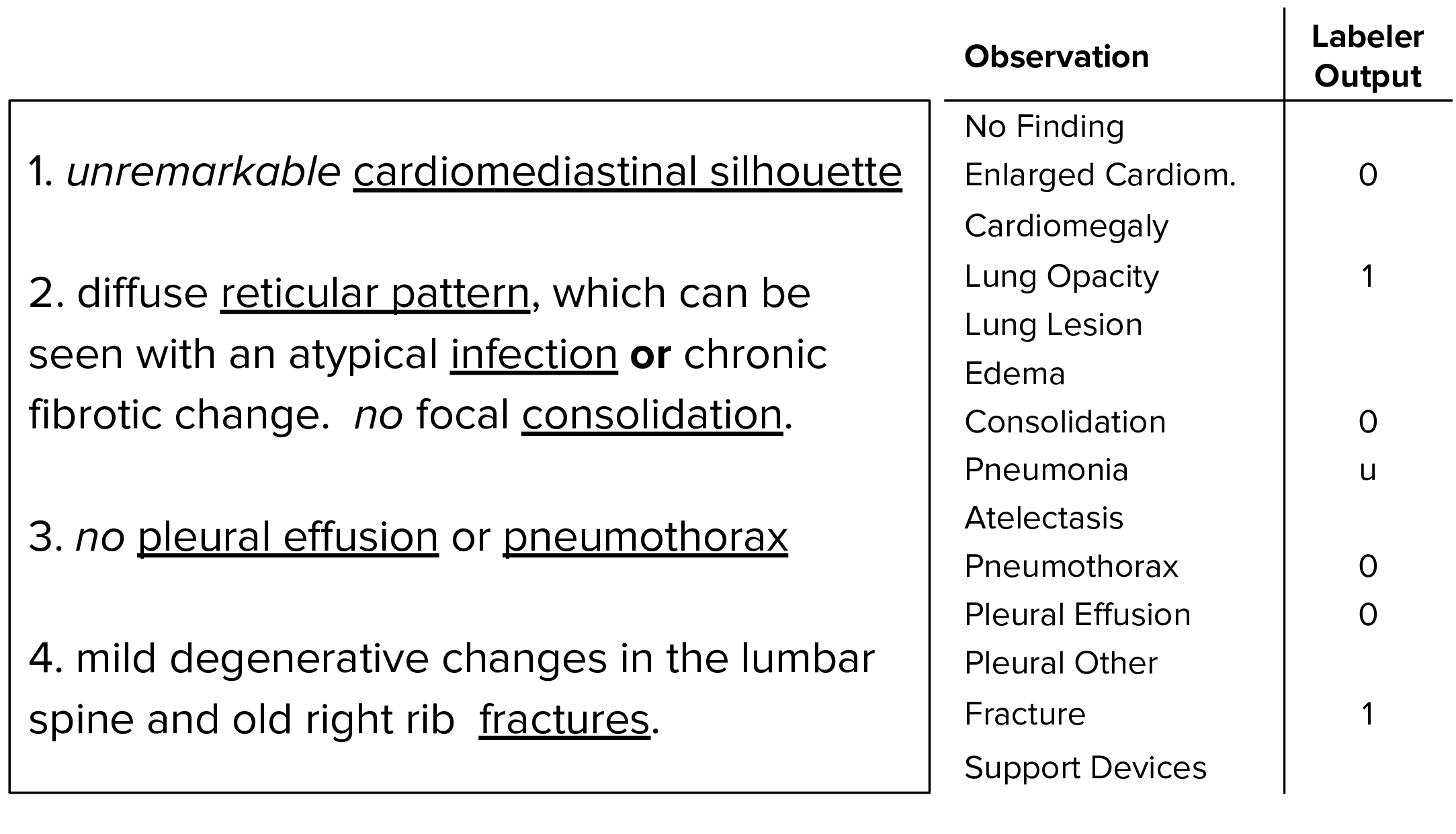
Our labeler is set up in three distinct stages: mention extraction, mention classification, and mention aggregation. In the mention extraction stage, the labeler extracts mentions from a list of observations from the Impression section of radiology reports, which summarizes the key findings in the radiographic study. In the mention classification stage, mentions of observations are classified as negative, uncertain, or positive. In the mention aggregation stage, we use the classification for each mention of observations to arrive at a final label for the 14 observations (blank for unmentioned, 0 for negative, -1 for uncertain, and 1 for positive).
Use the labeling tool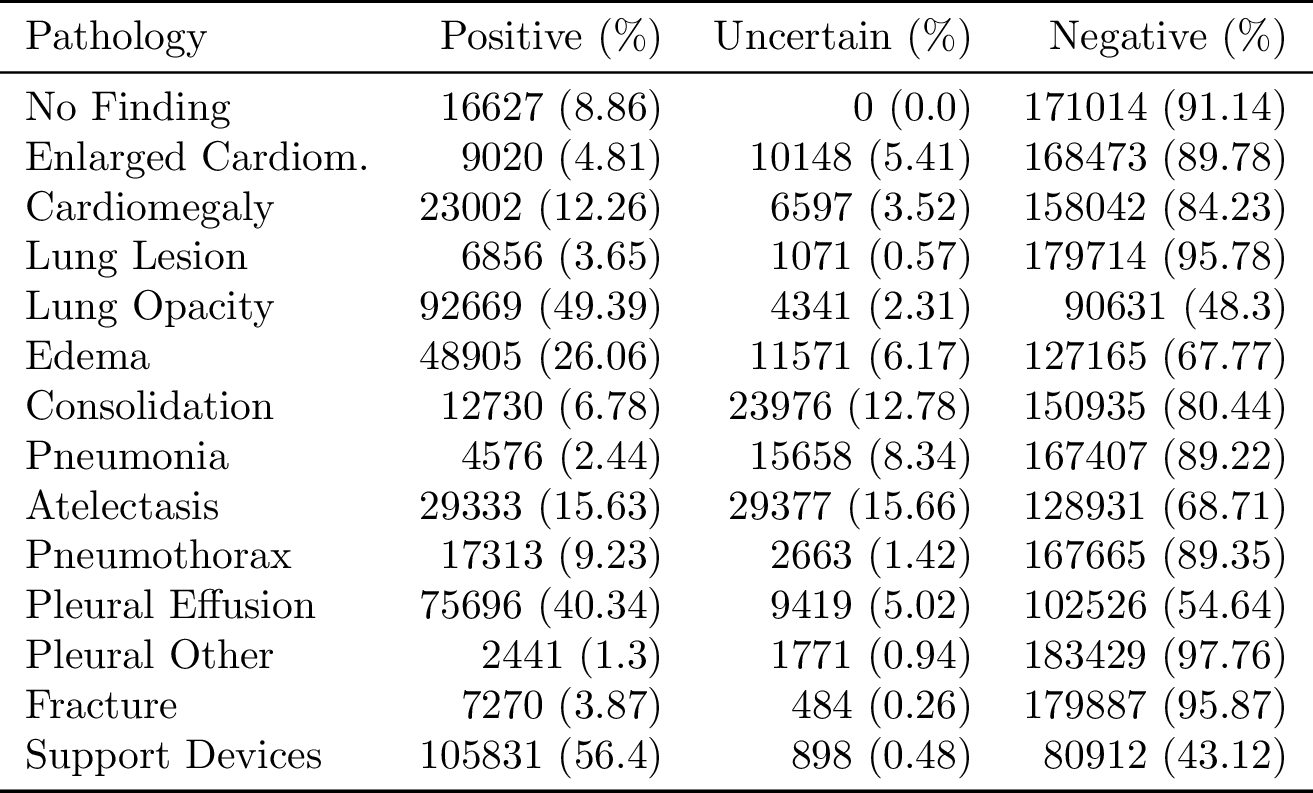
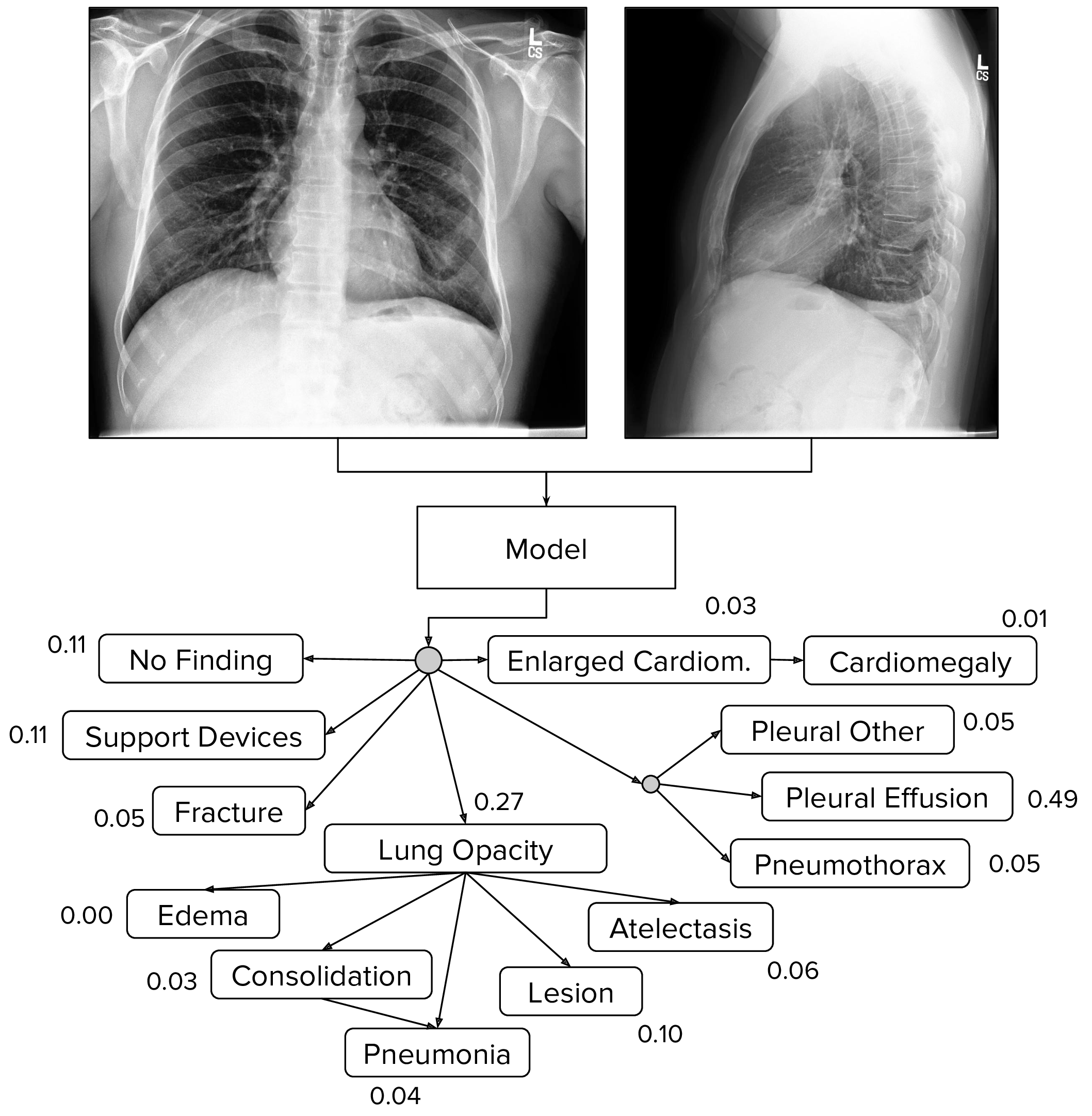
We train models that take as input a single-view chest radiograph and output the probability of each of the 14 observations. When more than one view is available, the models output the maximum probability of the observations across the views.
The training labels in the dataset for each observation are either 0 (negative), 1 (positive), or u (uncertain). We explore different approaches to using the uncertainty labels during the model training.
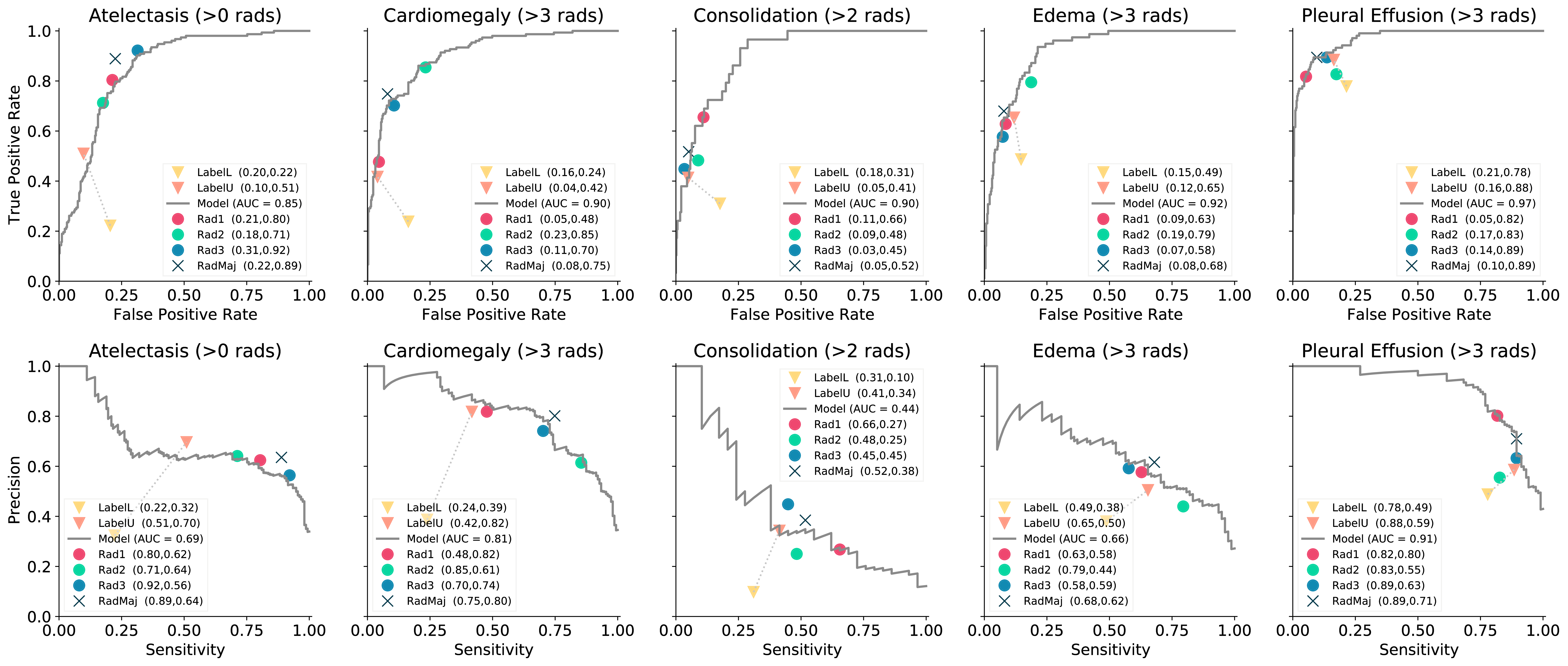
We focus on the evaluation of 5 observations which we call the competition tasks, selected based of clinical importance and prevalence: (a) Atelectasis, (b) Cardiomegaly, (c) Consolidation, (d) Edema, and (e) Pleural Effusion. We compare the performance of the different uncertainty approaches on a validation set of 200 studies on which the consensus of three radiologist annotations serves as ground truth. Our baseline model is selected based on the best performing approach on each competition tasks on the validation set: U-Ones for Atelectasis and Edema, U-MultiClass for Cardiomegaly and Pleural Effusion, and U-SelfTrained for Consolidation.
The test set consists of 500 studies from 500 unseen patients. Eight board-certified radiologists individually annotated each of the studies in the test set, classifying each observation into one of present, uncertain likely, uncertain unlikely, and absent. Their annotations were binarized such that all present and uncertain likely cases are treated as positive and all absent and uncertain unlikely cases are treated as negative. The majority vote of 5 radiologist annotations serves as a strong ground truth; the remaining 3 radiologist annotations were used to benchmark radiologist performance.
For each of the 3 individual radiologists and for their majority vote, we compute sensitivity (recall), specificity, and precision against the test set ground truth. To compare the model to radiologists, we plot the radiologist operating points with the model on both the ROC and Precision-Recall (PR) space. We examine whether the radiologist operating points lie below the curves to determine if the model is superior to the radiologists.
The model achieves the best AUC on Pleural Effusion (0.97), and the worst on Atelectasis (0.85). The AUC of all other observations are at least 0.9. On Cardiomegaly, Edema, and Pleural Effusion, the model achieves higher performance than all 3 radiologists but not their majority vote. On Consolidation, model performance exceeds 2 of the 3 radiologists, and on Atelectasis, all 3 radiologists perform better than the model.
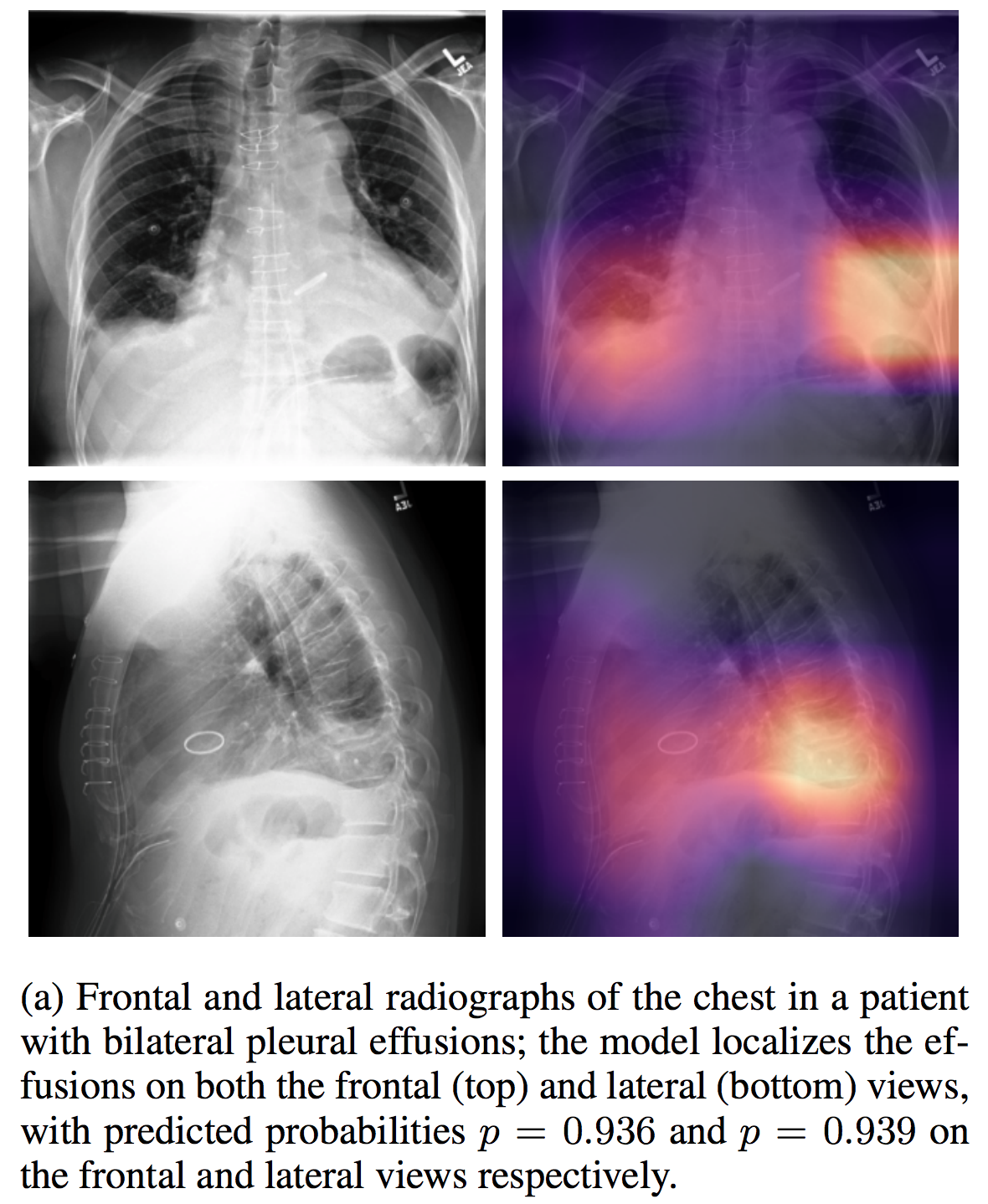
We're co-releasing our dataset with MIMIC-CXR, a large dataset of 371,920 chest x-rays associated with 227,943 imaging studies sourced from the Beth Israel Deaconess Medical Center between 2011 - 2016. Each imaging study can pertain to one or more images, but most often are associated with two images: a frontal view and a lateral view. Images are provided with 14 labels derived from a natural language processing tool applied to the corresponding free-text radiology reports.
Both our dataset and MIMIC-CXR share a common labeler, the CheXpert labeler, for deriving the same set of labels from free-text radiology reports.
One of the main obstacles in the development of chest radiograph interpretation models has been the lack of datasets with strong radiologist-annotated groundtruth and expert scores against which researchers can compare their models. We hope that CheXpert will address that gap, making it easy to track the progress of models over time on a clinically important task.
Furthermore, we have developed and open-sourced the CheXpert labeler, an automated rule-based labeler to extract observations from the free text radiology reports to be used as structured labels for the images. We hope that this makes it easy to help other institutions extract structured labels from their reports and release other large repositories of data that will allow for cross-institutional testing of medical imaging models.
Finally, we hope that the dataset will help development and validation of chest radiograph interpretation models towards improving healthcare access and delivery worldwide.

In the U.S., about half of all radiology studies are x-rays, mostly of the chest. Chest x-ray studies are even more common around the world. Chest x-ray interpretation is a “bread and butter” problem for radiologists with vital public health implications. Chest x-rays can stop the spread of tuberculosis, detect lung cancer early, and support the responsible use of antibiotics.

Ground truth is critical in evaluating deep learning models in medical imaging and provide the foundation for clinical relevance when interpreting results in this field - this is why we focus a lot of our effort on considering the best available ground truth via a panel of medical sub specialist experts to best understand the clinical implication of our model results.
Find the dataset on the Stanford AIMI website. The test set labels (and image links) are available on this GitHub repository.